This tutorial will show how you can start the FastRet GUI and explain the features it provides.
Starting the GUI
To start the GUI, install the package and then run the following command in an interactive R terminal:
FastRet::start_gui()After running the above code, you should see an output like
Listening on http://localhost:8080in your R console. This means that the GUI is now running and you can access it via the URL http://localhost:8080 in your browser. If your terminal supports it, you can also click on the displayed link.
By default, the GUI opens in mode Train new Model. To apply or adjust pretrained models, select mode Predict Retention Time or Adjust existing Model instead. For more information about the individual modes and the various input fields, click on the little question mark symbols next to the different input fields or read the following sections.
Train new Model
In mode Train new Model you can upload excel files containing the names, SMILES and retention times of metabolites measured on your specific chromatography column and use this data to train a predictive model. FastRet includes an example Excel file with retention times for 442 metabolites measured on a reverse phase liquid chromatography column at a temperature of 35 degree celsius and a flowrate of 0.3ml/min. To print the file path of this excel file and a preview of its contents, enter the following lines an interactive R session:
path <- system.file("extdata", "RP.xlsx", package = "FastRet")
cat(path, "\n", sep = "")
#> /home/runner/work/_temp/Library/FastRet/extdata/RP.xlsx
df <- openxlsx::read.xlsx(path, 1)
head(df)
#> RT SMILES
#> 1 3.34 CCC(C(=O)O)O
#> 2 3.35 COC1=C(C=CC(=C1)CCN)O
#> 3 2.11 C1=NC2=C(N1)C(=NC=N2)N
#> 4 2.10 C1=NC2=C(C(=N1)N)N=CN2C3C(C(C(O3)COP(=O)(O)O)O)O
#> 5 3.13 C1C2C(C(C(O2)N3C=NC4=C3N=CN=C4N)O)OP(=O)(O1)O
#> 6 2.07 C1=NC2=C(C(=N1)N)N=CN2C3C(C(C(O3)COP(=O)(O)O)OP(=O)(O)O)O
#> NAME
#> 1 2-HYDROXYBUTYRIC ACID
#> 2 3-METHOXYTYRAMINE
#> 3 ADENINE
#> 4 ADENOSINE 5'-MONOPHOSPHATE
#> 5 ADENOSINE 3',5'-CYCLIC MONOPHOSPHATE
#> 6 ADENOSINE 3',5'-DIPHOSPHATETo start model training, upload your Excel file and click the
Train Model button. Training the model might take some time,
depending on the size of the training set. When you click on
Show console logs you can see the progress of the training
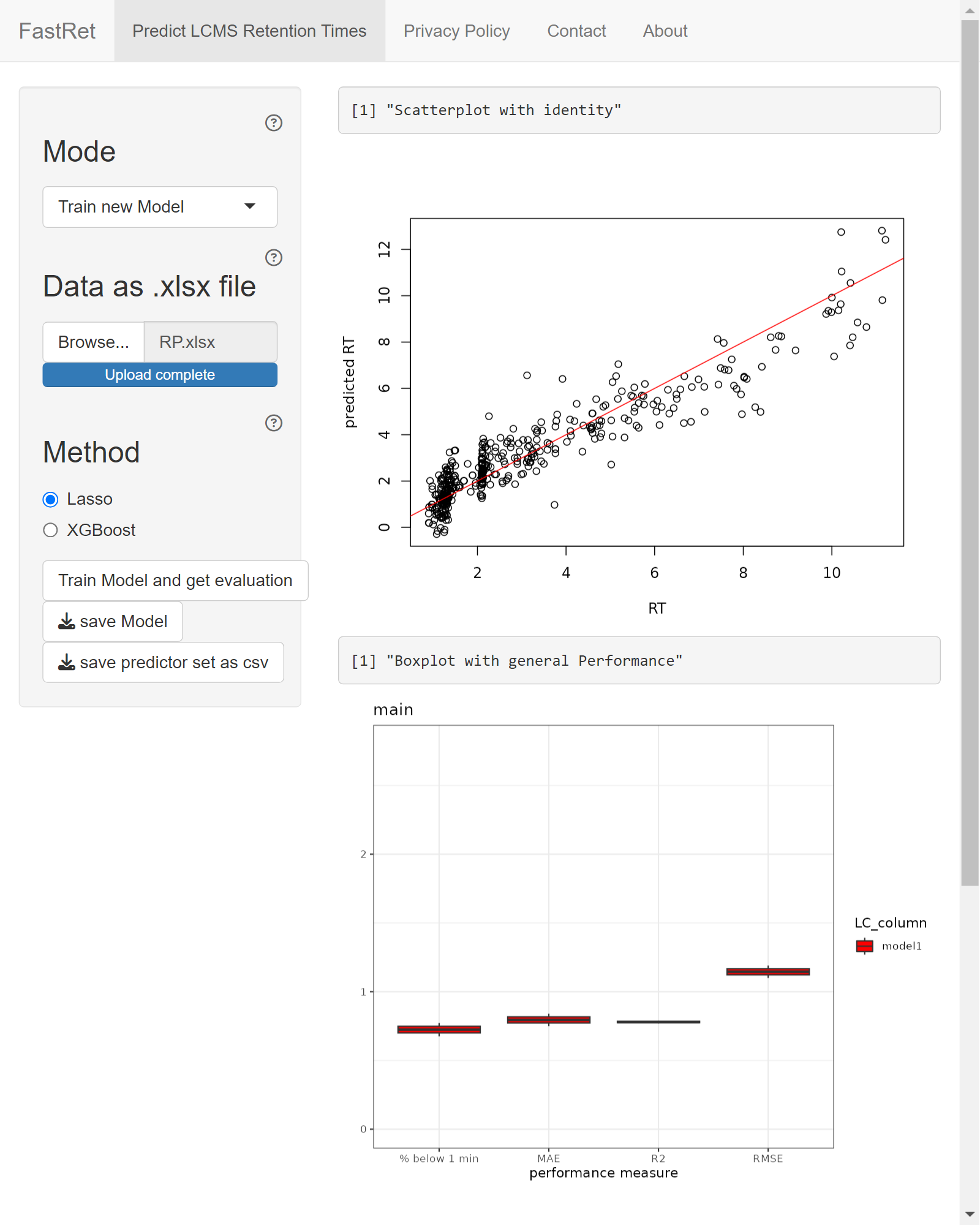
process. Upon completion, performance measures and a table of training
dataset is shown. For further details about the training process and the
performance measures, see section Model-Training
of article Package-Internals.
To use the trained model to predict retention times for new molecules, you have to:
- Save the model by clicking the Save Model button
- Switch to model Predict Retention Times
- Upload the model again while in mode Predict Retention Times
- Enter the SMILES of the molecules you want to predict retention times for
- Press Predict Retention Times
A more detailed guide on using the FastRet GUI for prediction is given in the next section Predict Retention Times.
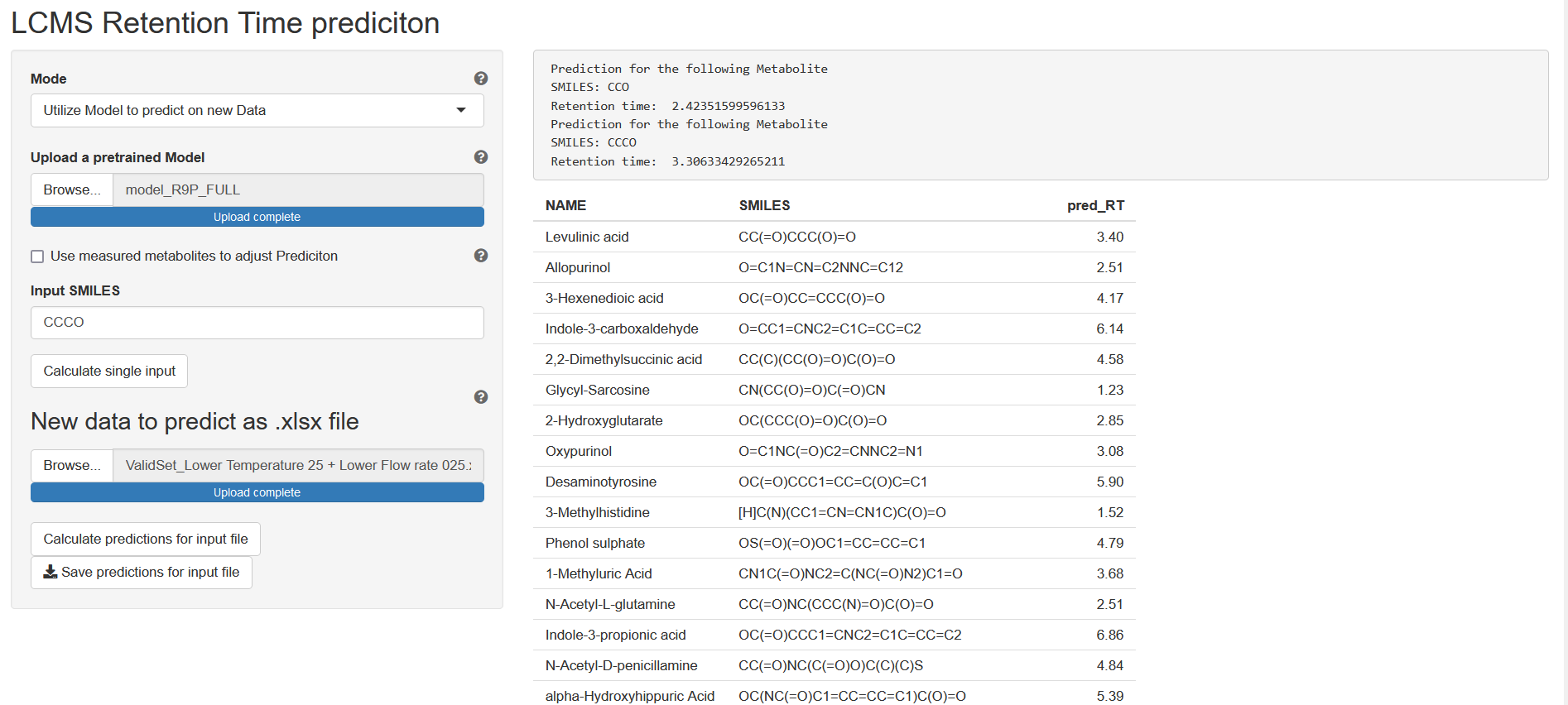
Predict Retention Times
In this mode, previously saved models can be used to make predictions for new data. To do so
- Click the Browse button from section Upload a pretrained Model and select the model you saved in the previous step
- Either enter the SMILES of your molecule of interest in the Input SMILES text field or
- Click the Browse button from section Upload SMILES as xlsx and select an Excel file containing columns NAME and SMILES
- Click button Predict at the bottom of the side bar
Adjusting existing model
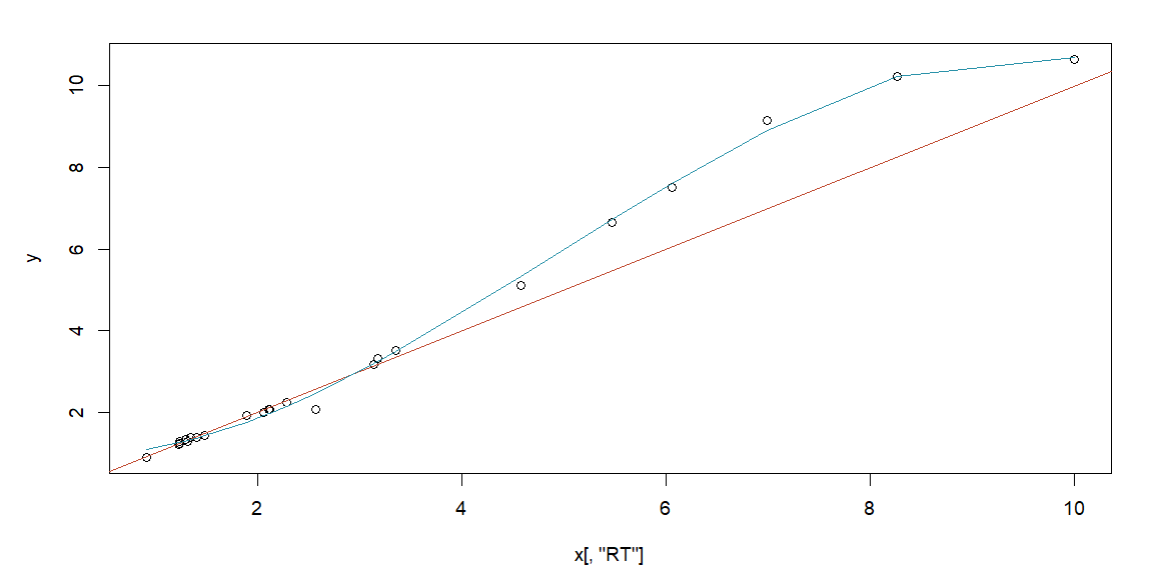
If you have measured some metabolites on your new experiment setup that were also measured on the original setup, you can use this method to adjust your model for your new column. To do so, switch to mode Adjust existing Model and upload the model you want to adjust. Then upload an Excel file containing the retention times of the metabolites measured on your new column. The Excel file should contain columns NAME, SMILES and RT. After clicking the Adjust Model button, the model will be adjusted and you can use it to predict retention times for new molecules measured on your new column.
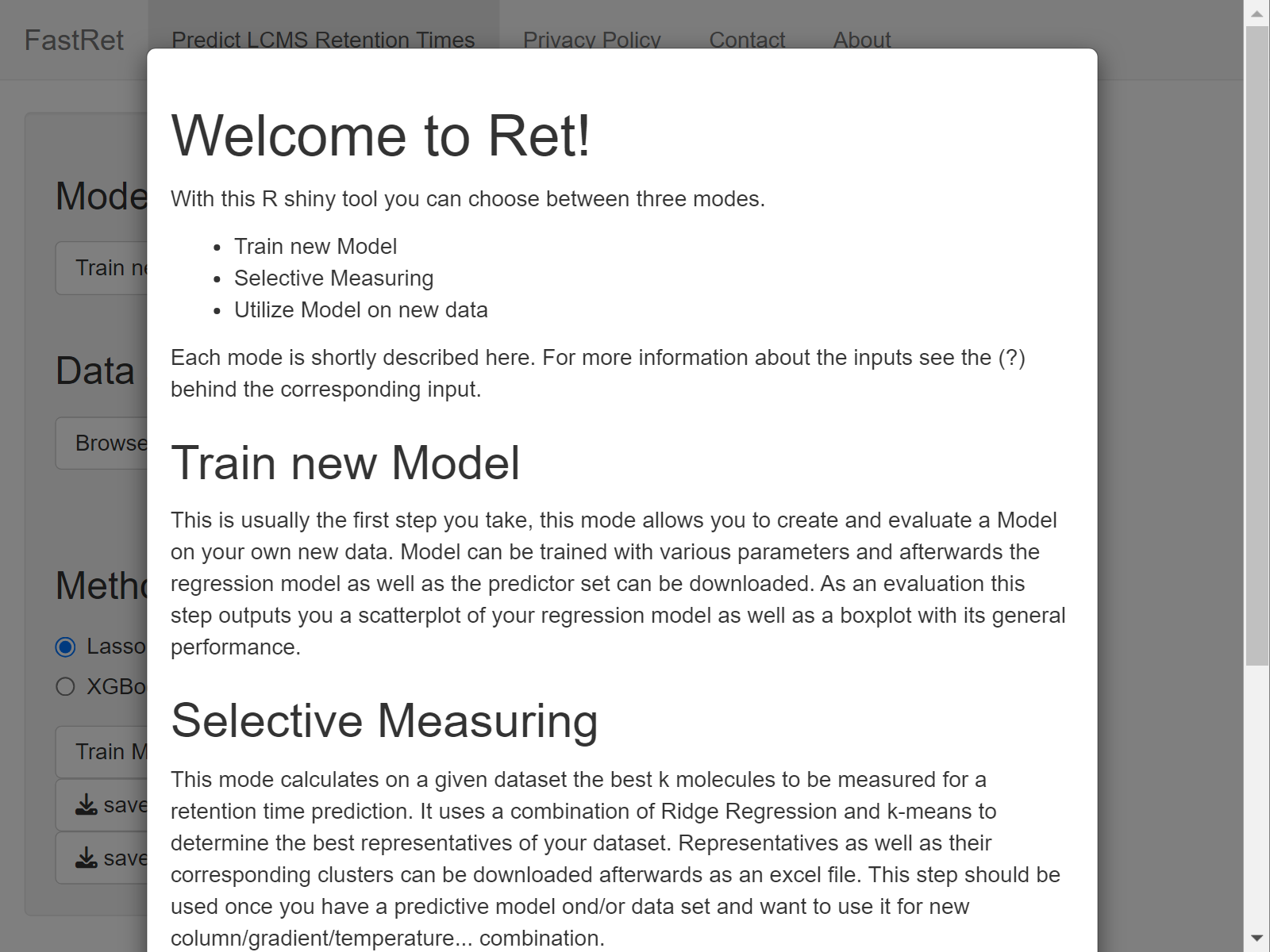
Selective Measuring
This mode calculates, for a given data set, the best k molecules to be measured for a retention time prediction on a new experiment setup. It uses a combination of Ridge Regression and k-means to determine the best representatives of your dataset. Representatives as well as their corresponding clusters can be downloaded afterwards as an excel file. This step should be used once you have a predictive model and/or data set and want to adjust it to work for a new column with adjusted chromatographic properties such as gradient, temperature, etc.