FAQ
Water Artifact
Question: When I choose n for
Water artefact fully inside red vertical lines? (y/n),
metabodecon asks me to
Choose another half width range (in ppm) for the water artefact,
instead of left and right borders. Why is that and what should I enter
here?
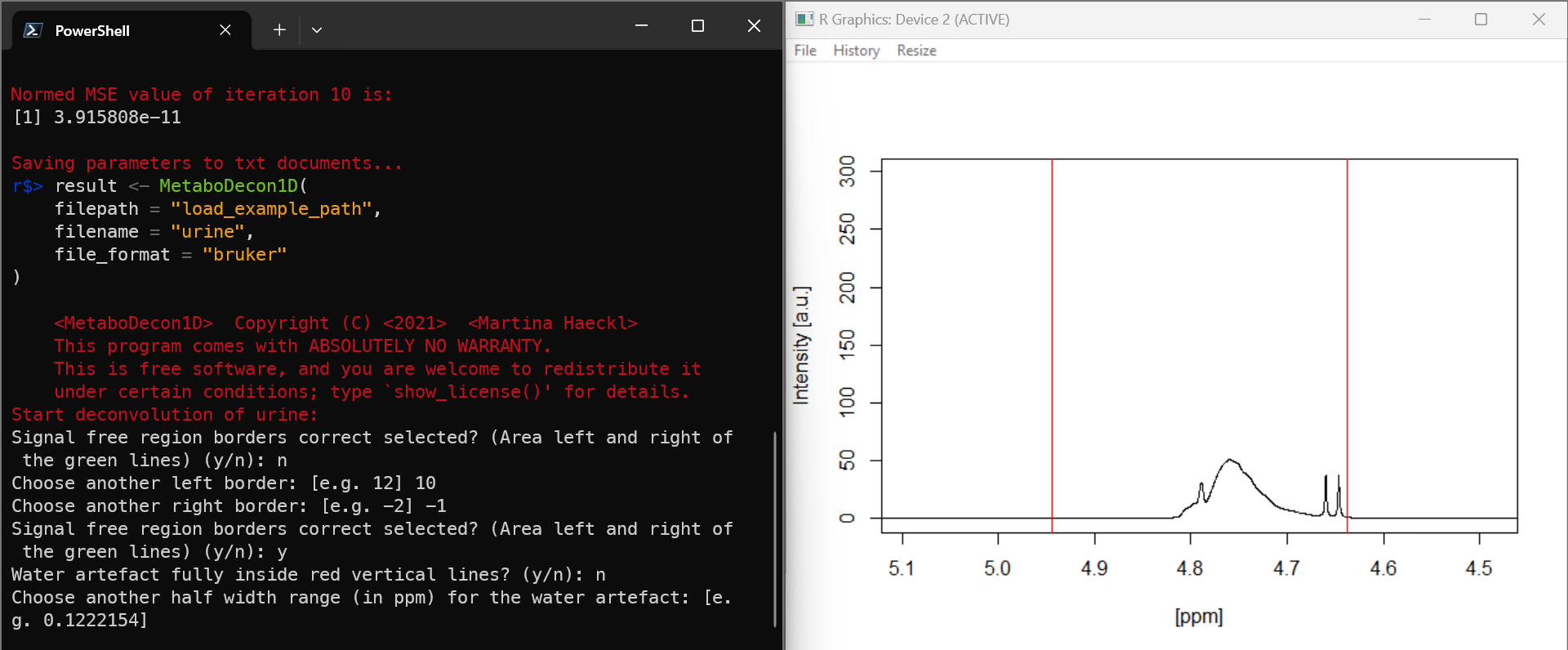
Answer: the water signal is always centered by Bruker, so you only have to specify the half width range to make the water area around the midpoint wider or narrower.
Parameter Optimization
Question: Why do you do exactly 10 iterations of parameter optimization?
Answer: This value was determined empirically. We found that 10 iterations are enough to get a good fit and more iterations would not improve the fit significantly, but would take longer. Obviously, a more objective stopping criterion would be better and will likely be implemented in a future version.
File Structure
Question: What file structure is expected for
bruker and jcampdx formats?
Answer: The expected file structure is as follows:
C:/bruker/urine # data_path (user input)
├── urine_1/ # name (user input)
│ └── 10/ # spectroscopy_value (user input), called expno in TopSpin manual
│ ├── acqus # acqus_file (constant)
│ └── pdata/ # processings_dir (constant)
│ └── 10/ # processing_value (user input), called procno in TopSpin manual
│ ├── 1r # spec_file (constant)
│ └── procs # spectrum_file (constant)
├── urine_2/...
└── ...
C:/jcampdx/urine # data_path (user input)
├── urine_1.dx # spectrum_file (user input)
├── urine_2.dx
└── ...