MetaboDecon1D Usage Example
The MetaboDecon1D package (v0.2.2) is the predecessor of
this package and can be downloaded from uni-regensburg.de/medicine/functional-genomics/staff/prof-wolfram-gronwald/software.
We include this usage example in here so we can easily reference old
behaviour and point out added features in metabodecon
(v1.x).
Install the package
- Open the following link in your browser: uni-regensburg.de/medicine/functional-genomics/staff/prof-wolfram-gronwald/software
- Click MetaboDecon1D: An R-package for the Deconvolution and Integration of 1D NMR data
- Download MetaboDecon1D_0.2.2.tar.gz
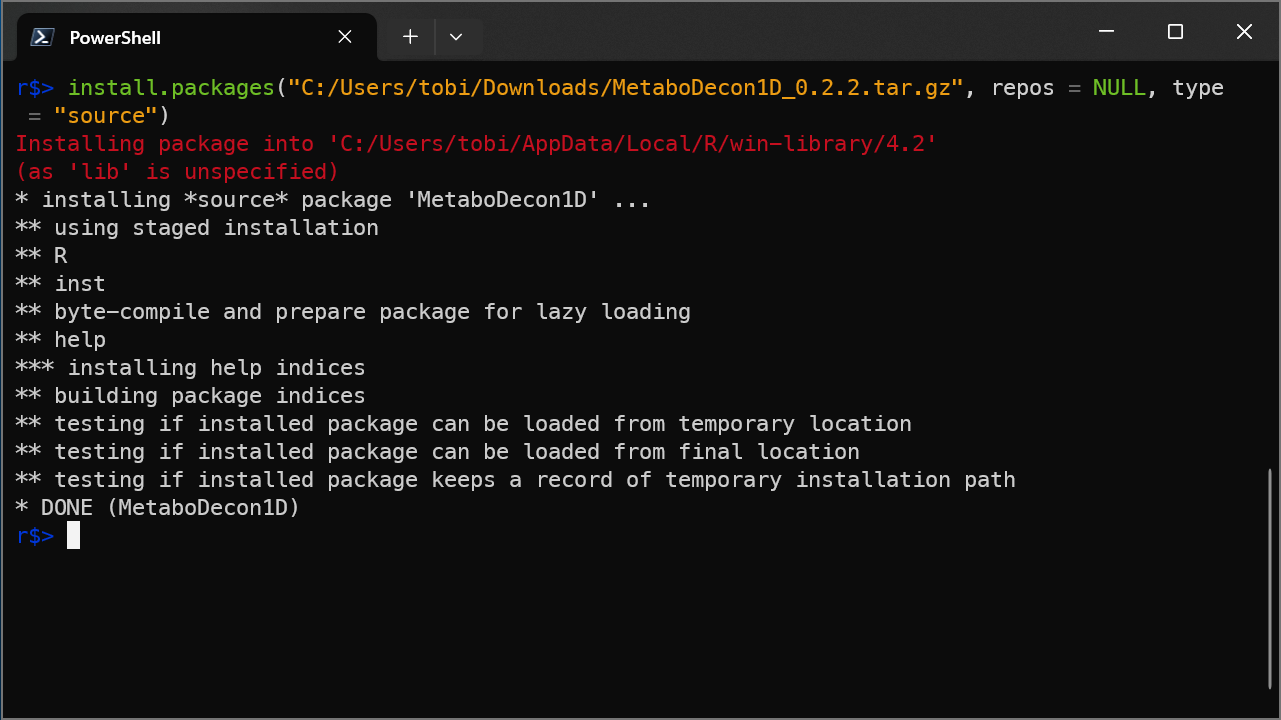
- Start an R session and enter command
install.packages("C:/Users/tobi/Downloads/MetaboDecon1D_0.2.2.tar.gz", repos = NULL, type = "source")(replaceC:/Users/tobi/Downloads/MetaboDecon1D_0.2.2.tar.gzwith the path to the downloaded file on your computer)
Load package
library(MetaboDecon1D)Deconvolute one spectrum in Bruker format
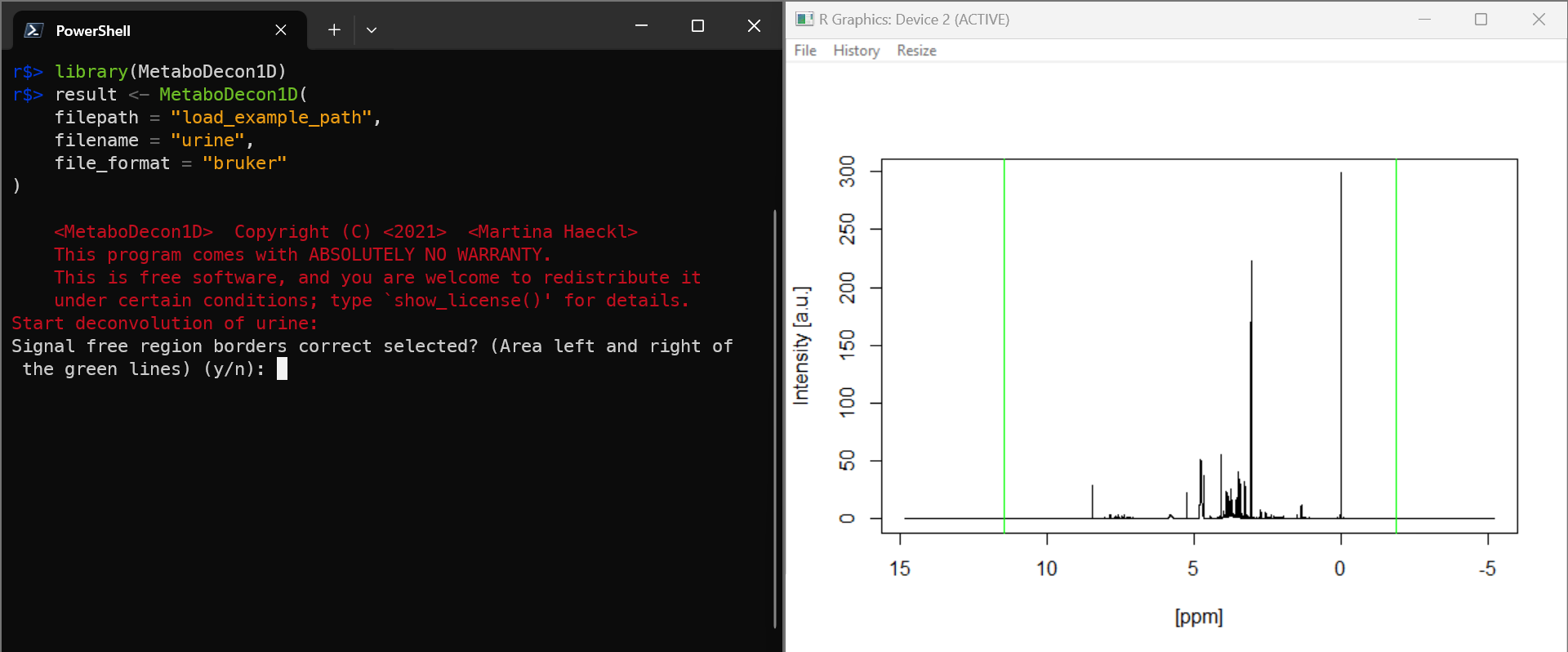
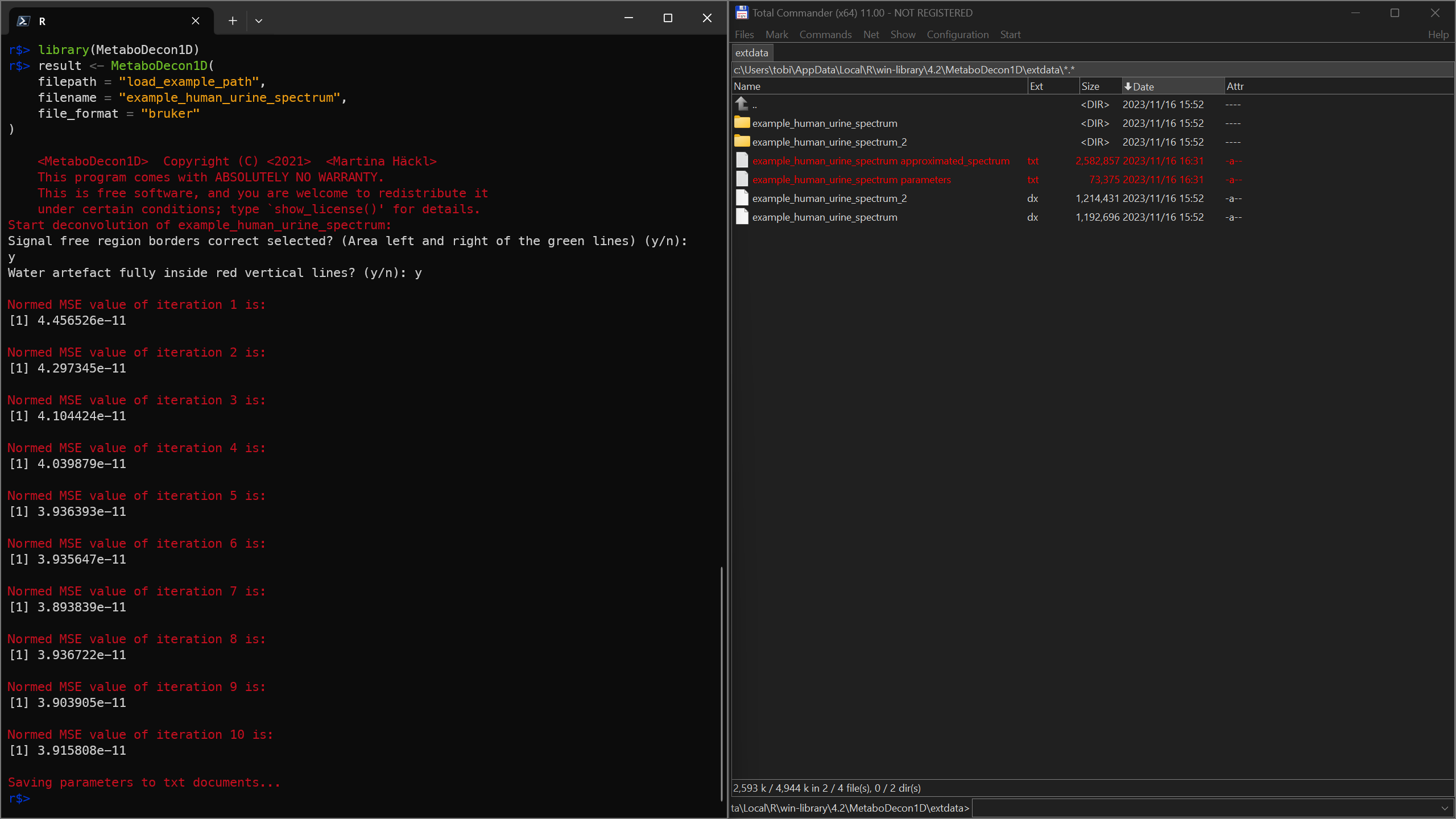
result <- MetaboDecon1D(
filepath = "load_example_path",
filename = "example_human_urine_spectrum",
file_format = "bruker"
)
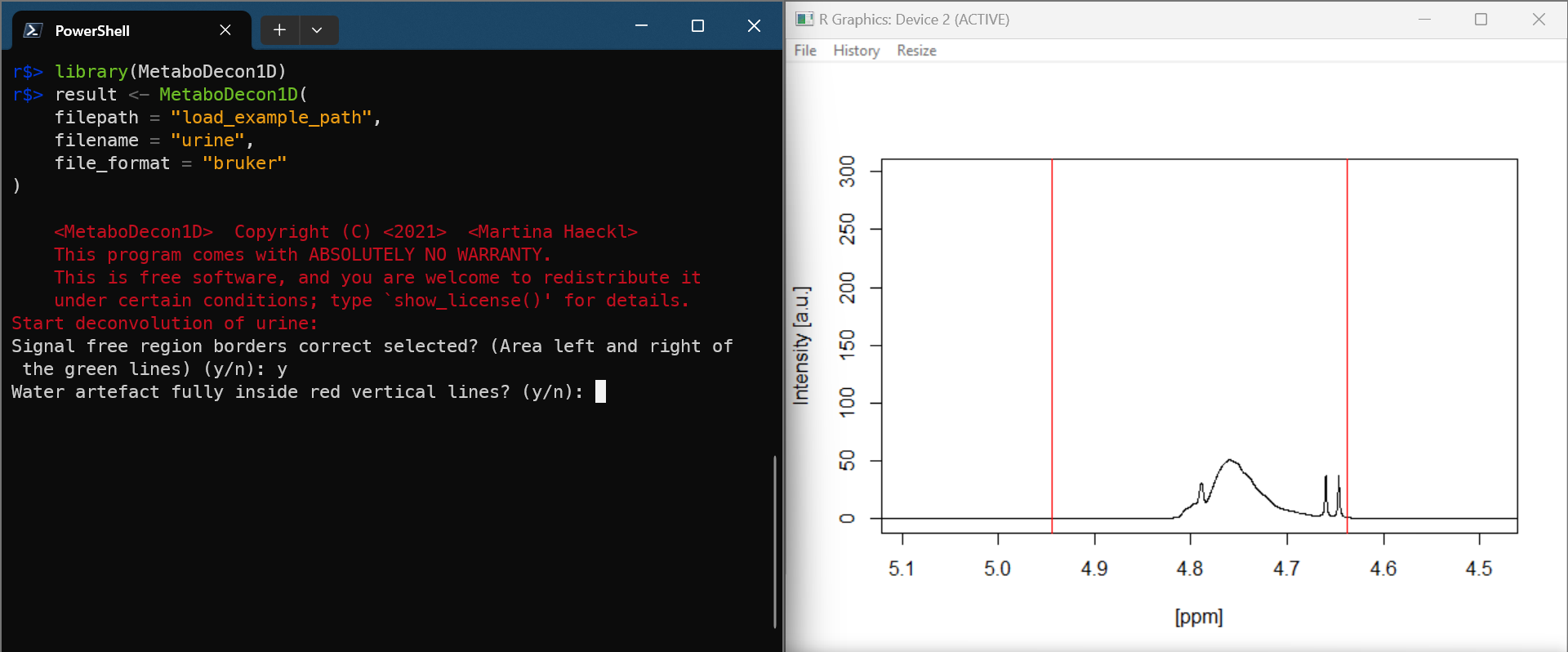
Visualize results and store plots as png
str(result)
plot_triplets(result)
plot_lorentz_curves_save_as_png(result)
plot_spectrum_superposition_save_as_png(result)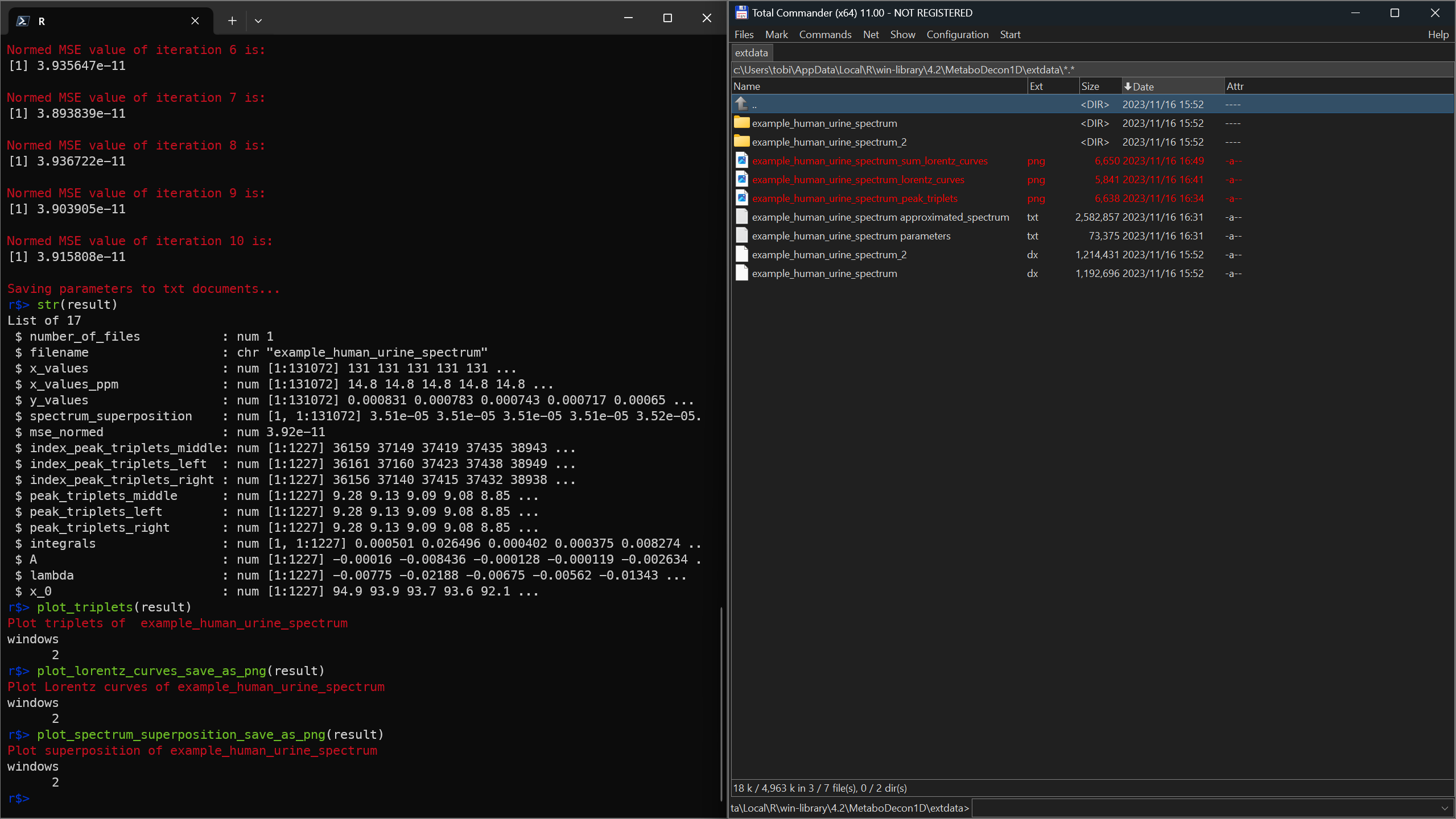
4_plots.png
Deconvolute one spectrum in jcampdx format
result <- MetaboDecon1D(
filepath = "load_example_path",
filename = "example_human_urine_spectrum.dx",
file_format = "jcampdx"
)
str(result)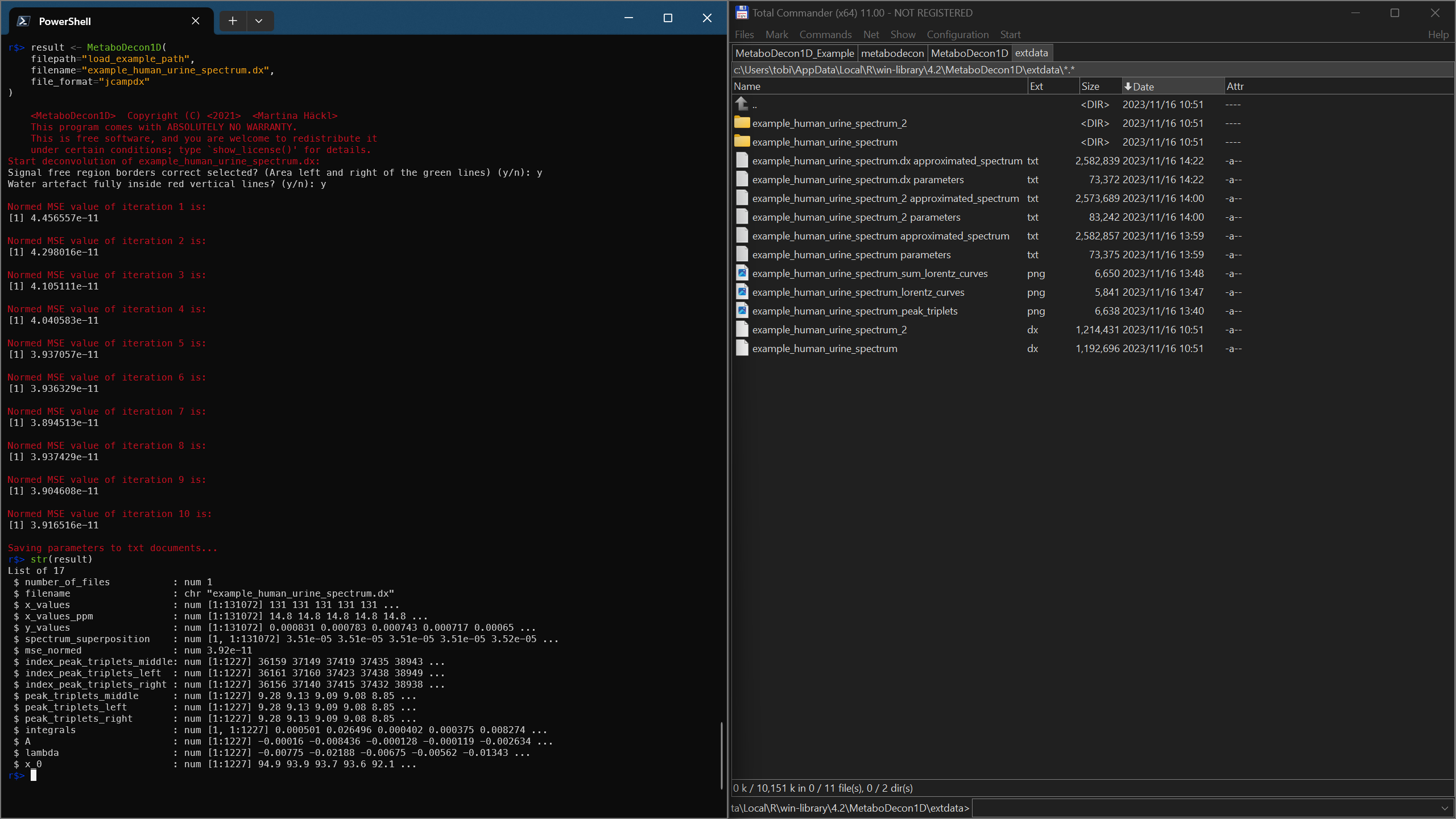
Deconvolute multiple spectra in Bruker format
result <- MetaboDecon1D(
filepath = "load_example_path",
file_format = "bruker"
)
str(result)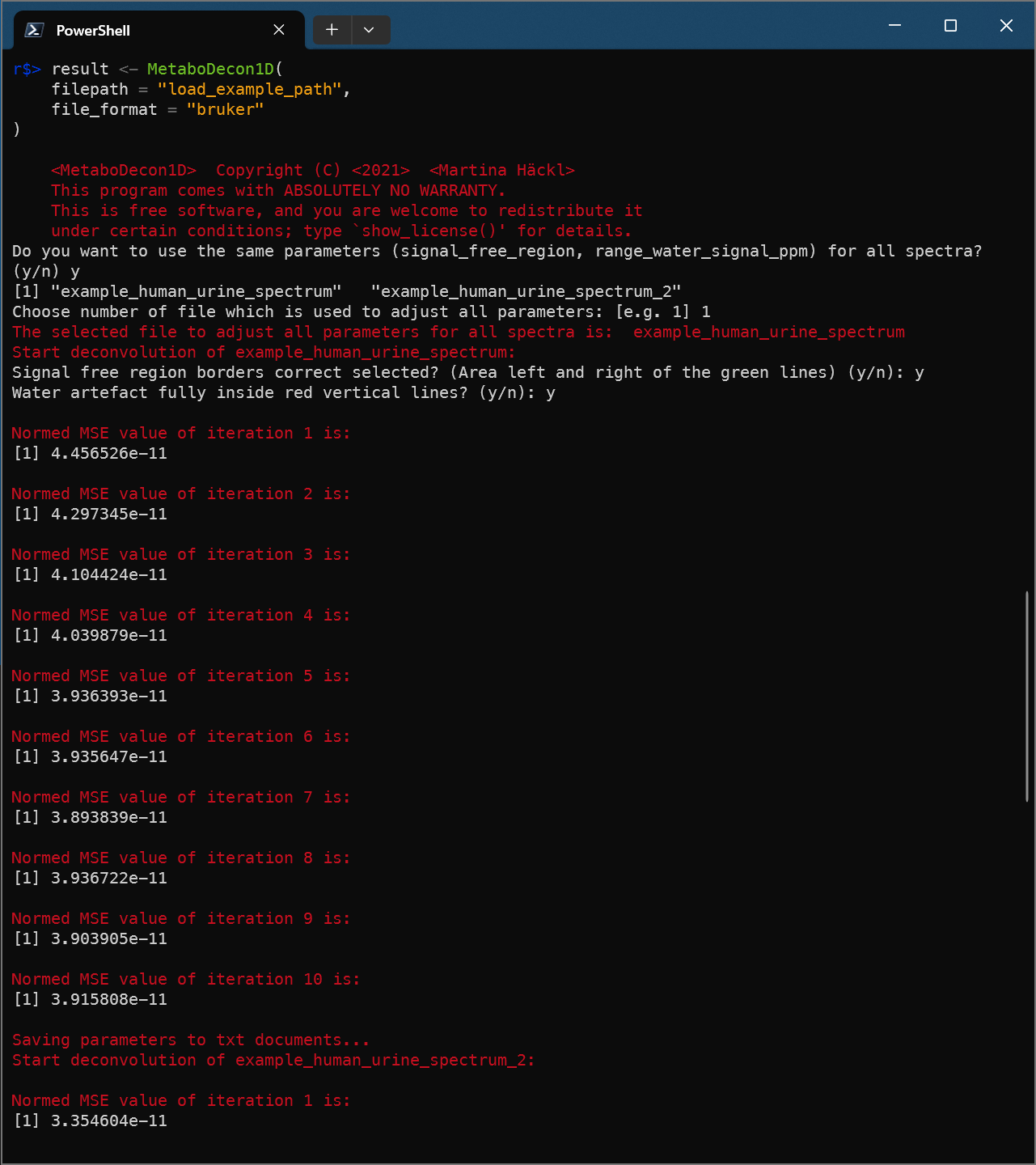
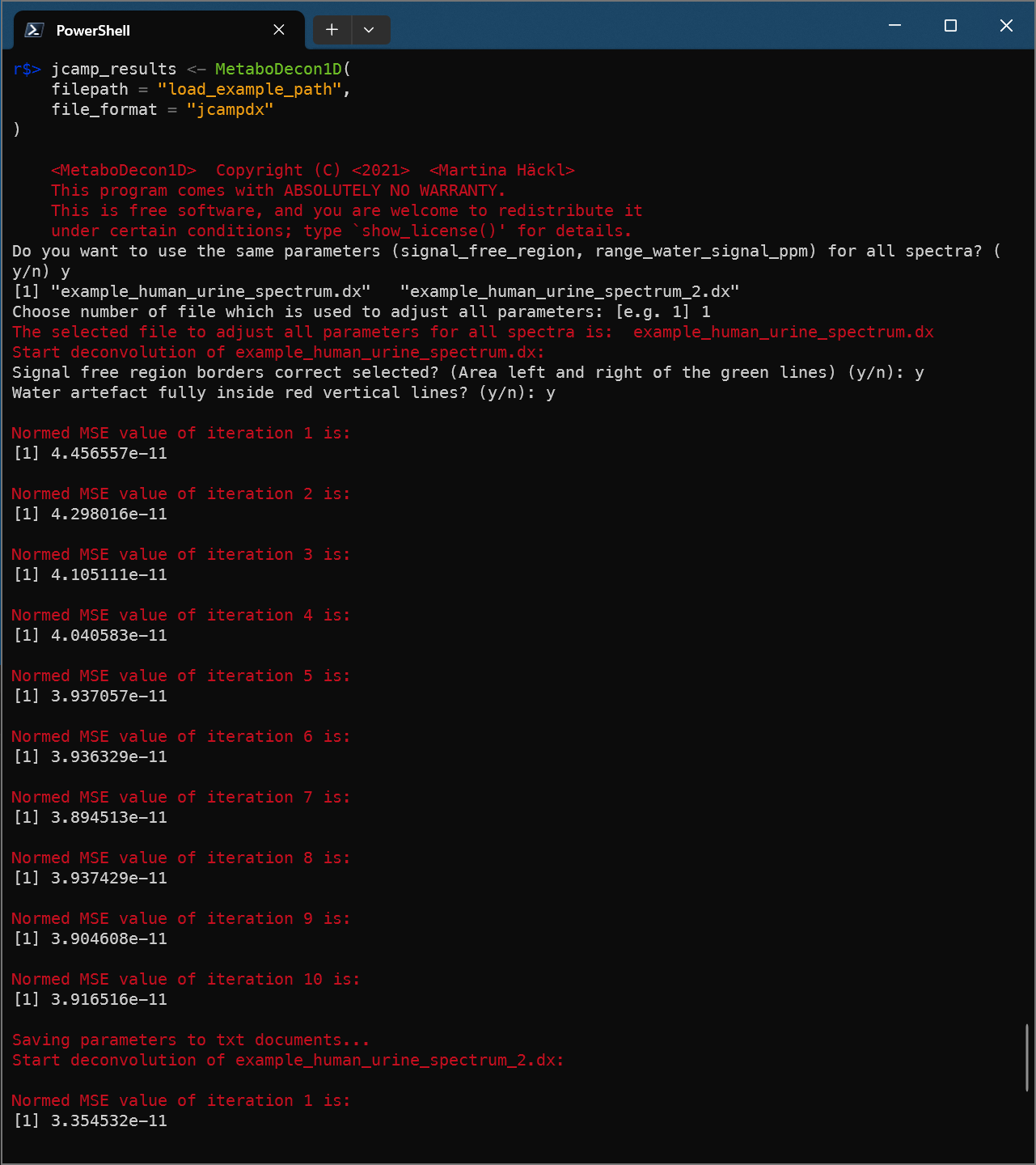
Deconvolute multiple spectra in jcampdx format
jcamp_results <- MetaboDecon1D(
filepath = "load_example_path",
file_format = "jcampdx"
)
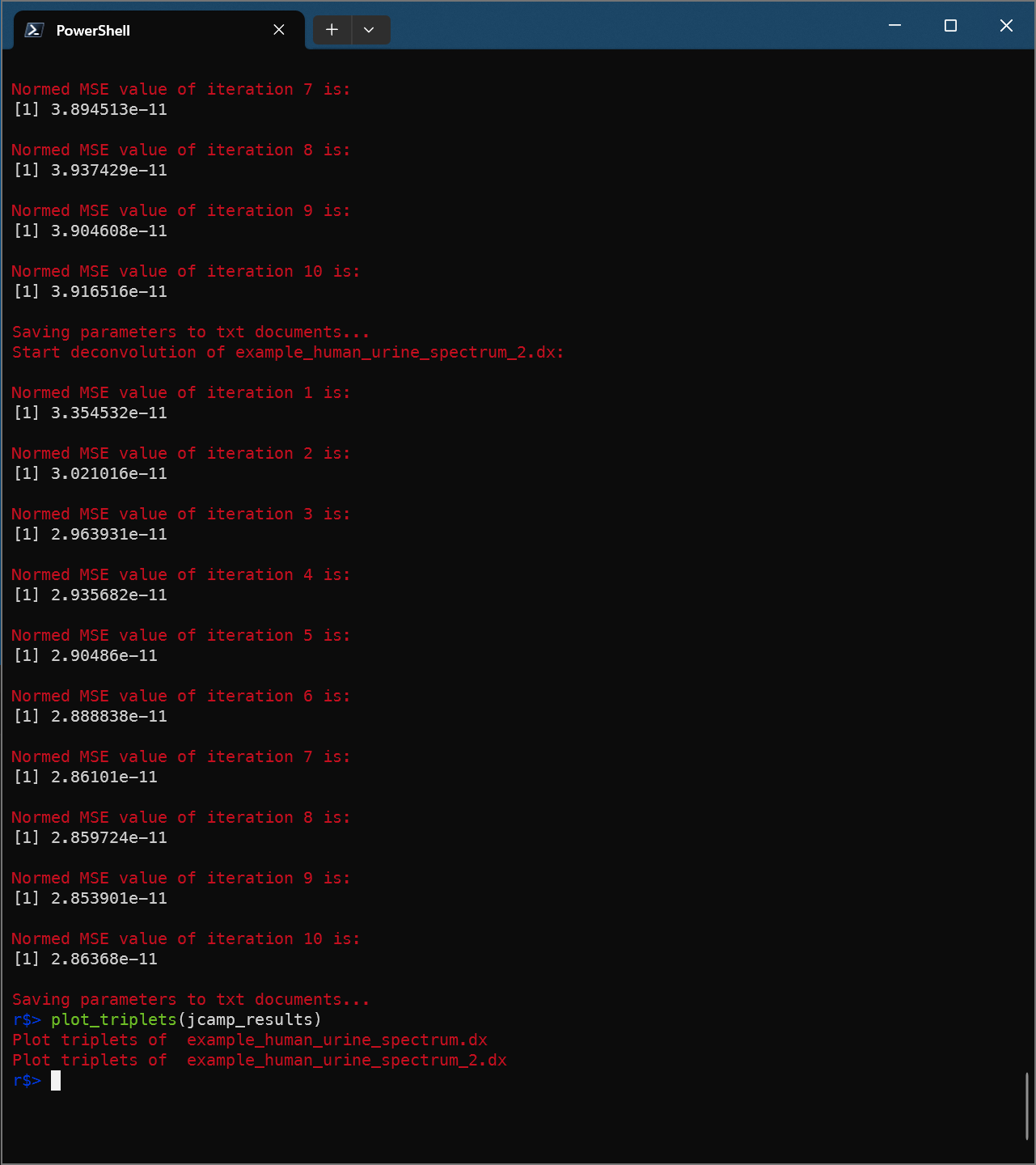
plot_triplets(jcamp_results)
plot_lorentz_curves_save_as_png(jcamp_results)
plot_spectrum_superposition_save_as_png(jcamp_results)